This function creates a matrix of individual density plots (i.e., a ridgeline matrix) or boxplots (for metric variables) or of individual barplots (for categorical variables). The age, period or cohort information can each either be plotted on the x-axis or the y-axis.
Usage
plot_densityMatrix(
dat,
y_var,
dimensions = c("period", "age"),
age_groups = NULL,
period_groups = NULL,
cohort_groups = NULL,
plot_type = "density",
highlight_diagonals = NULL,
y_var_cat_breaks = NULL,
y_var_cat_labels = NULL,
weights_var = NULL,
log_scale = FALSE,
legend_title = NULL,
...
)Arguments
- dat
Dataset with columns
periodandageand the main variable specified through argumenty_var.- y_var
Character name of the main variable to be plotted.
- dimensions
Character vector specifying the two APC dimensions that should be visualized along the x-axis and y-axis. Defaults to
c("period","age").- age_groups, period_groups, cohort_groups
Each a list. Either containing purely scalar values or with each element specifying the two borders of one row or column in the density matrix. E.g., if the period should be visualized in decade columns from 1980 to 2009, specify
period_groups = list(c(1980,1989), c(1990,1999), c(2000,2009)). The list can be named to specify labels for the categories. Only the two arguments must be passed that were specified by thedimensionsargument.- plot_type
One of
c("density","boxplot"). Only used if they_varcolumn is metric.- highlight_diagonals
Optional list to define diagonals in the density that should be highlighted with different colors. Each list element should be a numeric vector stating the index of the diagonals (counted from the top left) that should be highlighted in the same color. If the list is named, the names are used as legend labels.
- y_var_cat_breaks
Optional numeric vector of breaks to categorize
y_varbased on calling functioncut. Only used to highlight the categories based on different colors. And only used if they_varcolumn is numeric.- y_var_cat_labels
Optional character vector for the names of the categories that were defined based on
y_var_cat_breaks. The length of this vector must be one shorter thanlength(y_var_cat_breaks). Only used if they_varcolumn is numeric.- weights_var
Optional character name of a weights variable used to project the results in the sample to some population.
- log_scale
Indicator if the main variable should be log10 transformed. Only used if the
y_varcolumn is numeric. Defaults to FALSE.- legend_title
Optional plot annotation.
- ...
Additional arguments passed to
plot_density.
References
Weigert, M., Bauer, A., Gernert, J., Karl, M., Nalmpatian, A., Küchenhoff, H., and Schmude, J. (2021). Semiparametric APC analysis of destination choice patterns: Using generalized additive models to quantify the impact of age, period, and cohort on travel distances. Tourism Economics. doi:10.1177/1354816620987198.
Author
Alexander Bauer alexander.bauer@stat.uni-muenchen.de, Maximilian Weigert maximilian.weigert@stat.uni-muenchen.de
Examples
library(APCtools)
# define categorizations for the main trip distance
dist_cat_breaks <- c(1,500,1000,2000,6000,100000)
dist_cat_labels <- c("< 500 km","500 - 1,000 km", "1,000 - 2,000 km",
"2,000 - 6,000 km", "> 6,000 km")
age_groups <- list(c(80,89),c(70,79),c(60,69),c(50,59),c(40,49),c(30,39),c(20,29))
period_groups <- list(c(1970,1979),c(1980,1989),c(1990,1999),c(2000,2009),c(2010,2019))
cohort_groups <- list(c(1980,1989),c(1970,1979),c(1960,1969),c(1950,1959),c(1940,1949),
c(1930,1939),c(1920,1929))
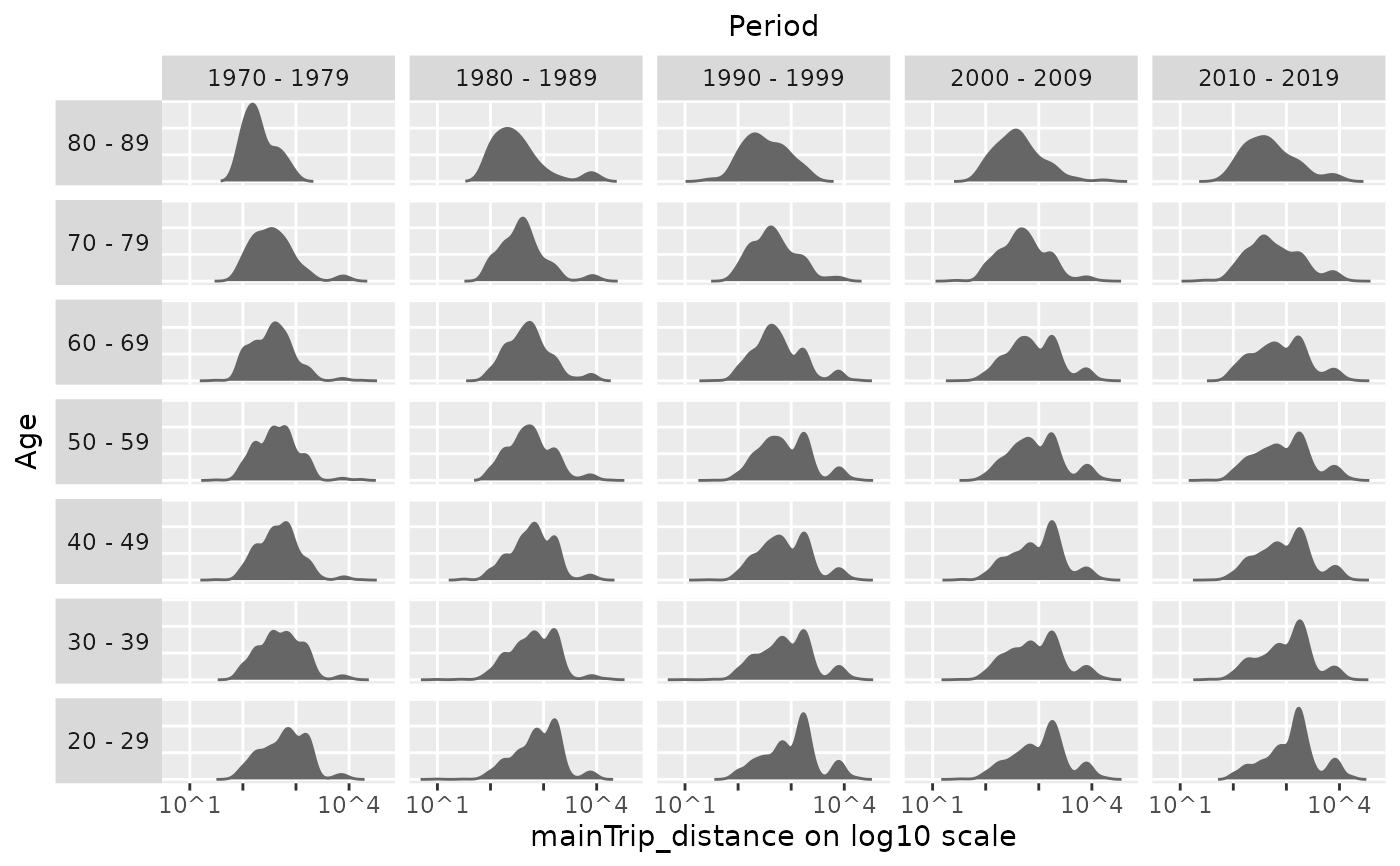
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
age_groups = age_groups,
period_groups = period_groups,
log_scale = TRUE)
#> Excluding 9149 missing observations of mainTrip_distance...
 # \donttest{
# highlight two cohorts
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
age_groups = age_groups,
period_groups = period_groups,
highlight_diagonals = list(8, 10),
log_scale = TRUE)
#> Excluding 9149 missing observations of mainTrip_distance...
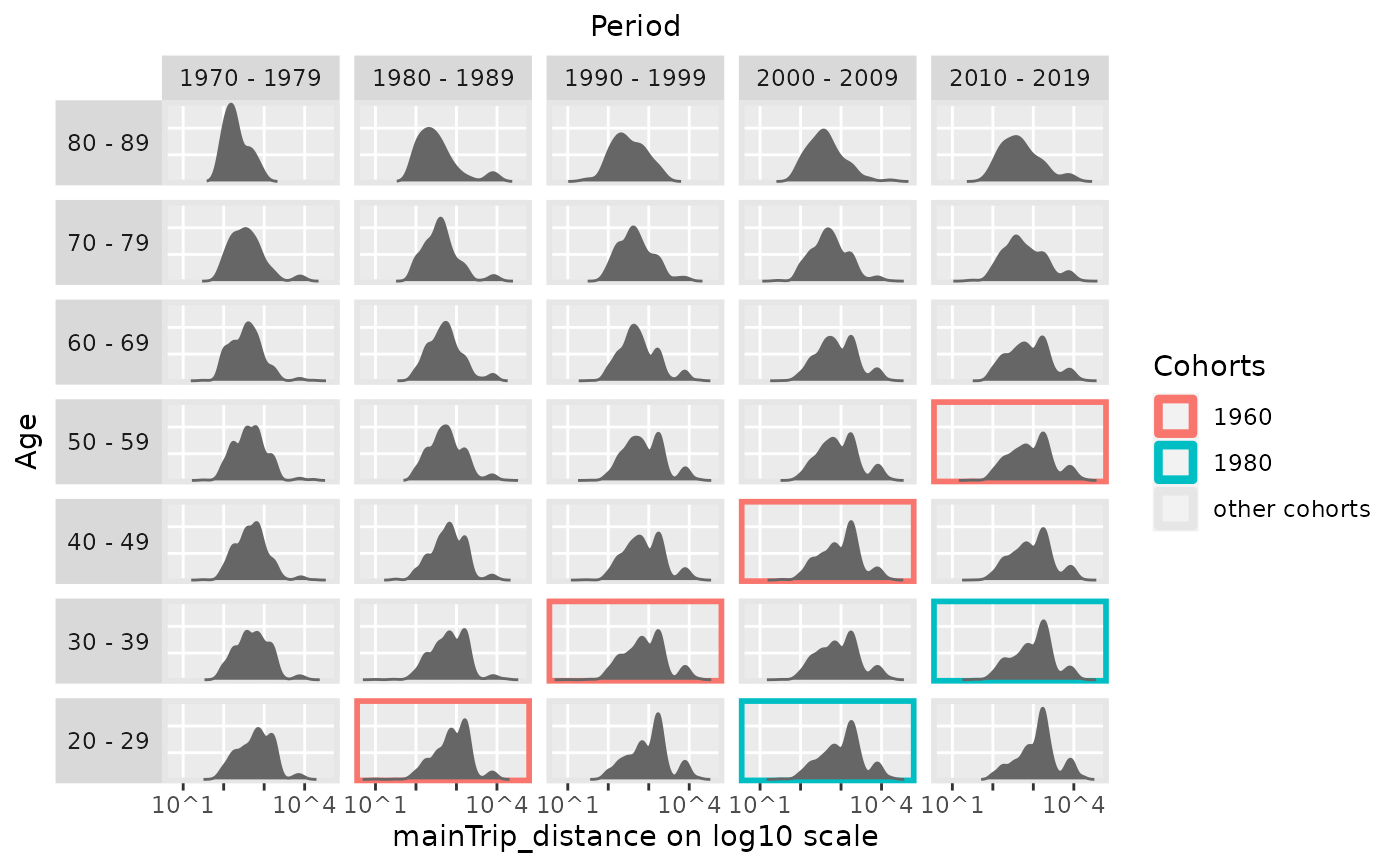
# \donttest{
# highlight two cohorts
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
age_groups = age_groups,
period_groups = period_groups,
highlight_diagonals = list(8, 10),
log_scale = TRUE)
#> Excluding 9149 missing observations of mainTrip_distance...
 # also mark different distance categories
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
age_groups = age_groups,
period_groups = period_groups,
log_scale = TRUE,
y_var_cat_breaks = dist_cat_breaks,
y_var_cat_labels = dist_cat_labels,
highlight_diagonals = list(8, 10),
legend_title = "Distance category")
#> Excluding 9149 missing observations of mainTrip_distance...
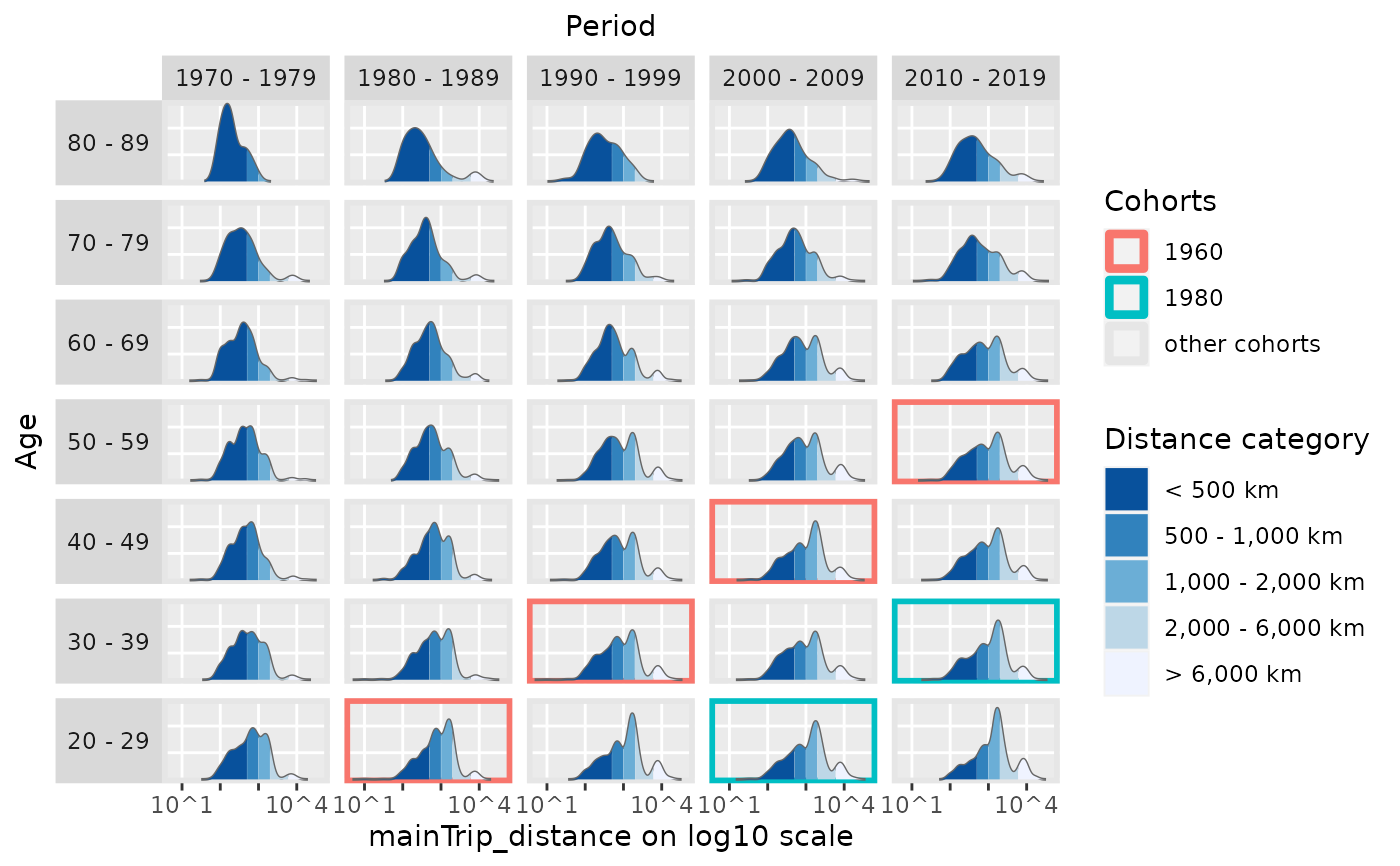
# also mark different distance categories
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
age_groups = age_groups,
period_groups = period_groups,
log_scale = TRUE,
y_var_cat_breaks = dist_cat_breaks,
y_var_cat_labels = dist_cat_labels,
highlight_diagonals = list(8, 10),
legend_title = "Distance category")
#> Excluding 9149 missing observations of mainTrip_distance...
 # flexibly assign the APC dimensions to the x-axis and y-axis
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
dimensions = c("period","cohort"),
period_groups = period_groups,
cohort_groups = cohort_groups,
log_scale = TRUE,
y_var_cat_breaks = dist_cat_breaks,
y_var_cat_labels = dist_cat_labels,
legend_title = "Distance category")
#> Excluding 7922 missing observations of mainTrip_distance...
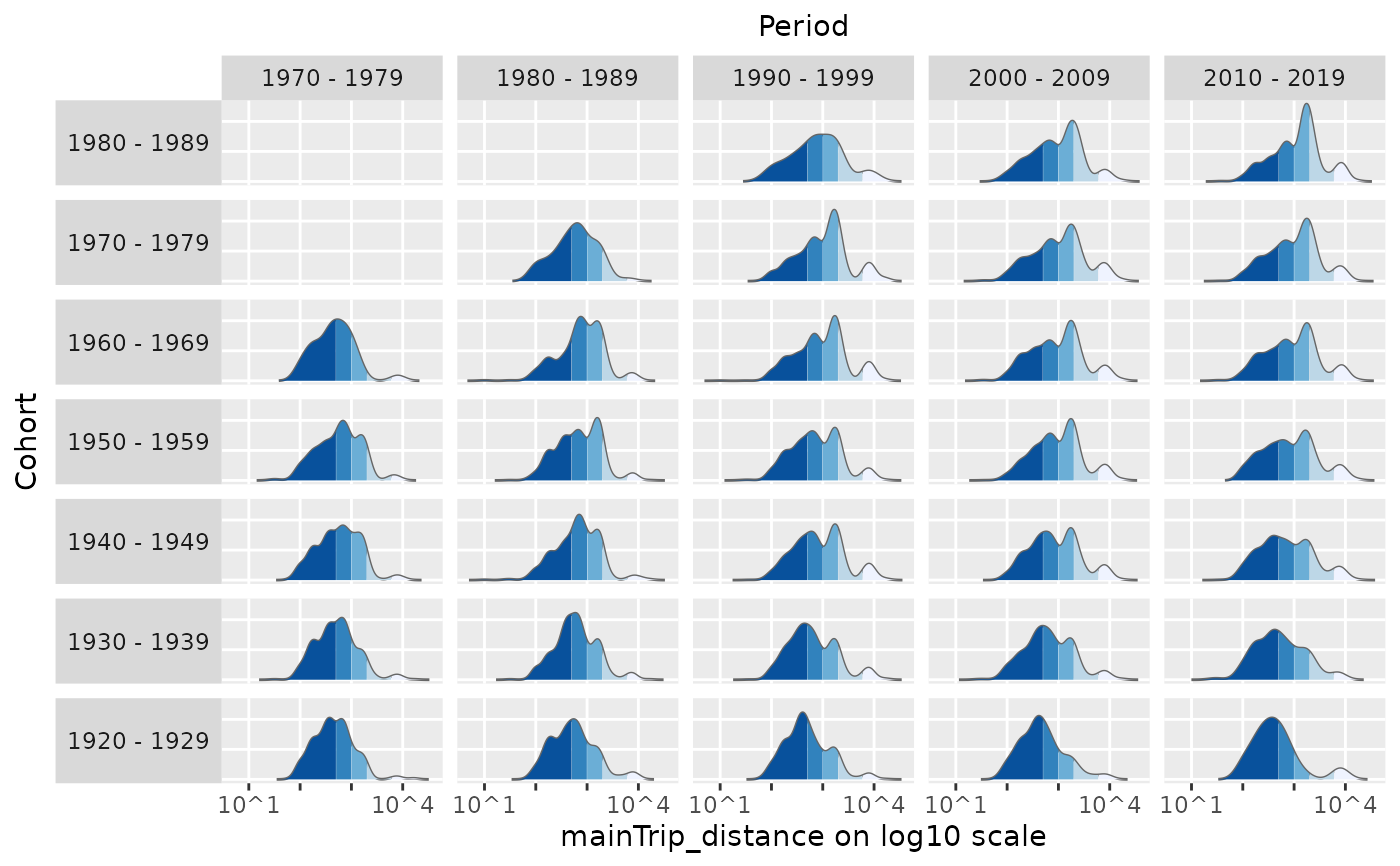
# flexibly assign the APC dimensions to the x-axis and y-axis
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
dimensions = c("period","cohort"),
period_groups = period_groups,
cohort_groups = cohort_groups,
log_scale = TRUE,
y_var_cat_breaks = dist_cat_breaks,
y_var_cat_labels = dist_cat_labels,
legend_title = "Distance category")
#> Excluding 7922 missing observations of mainTrip_distance...
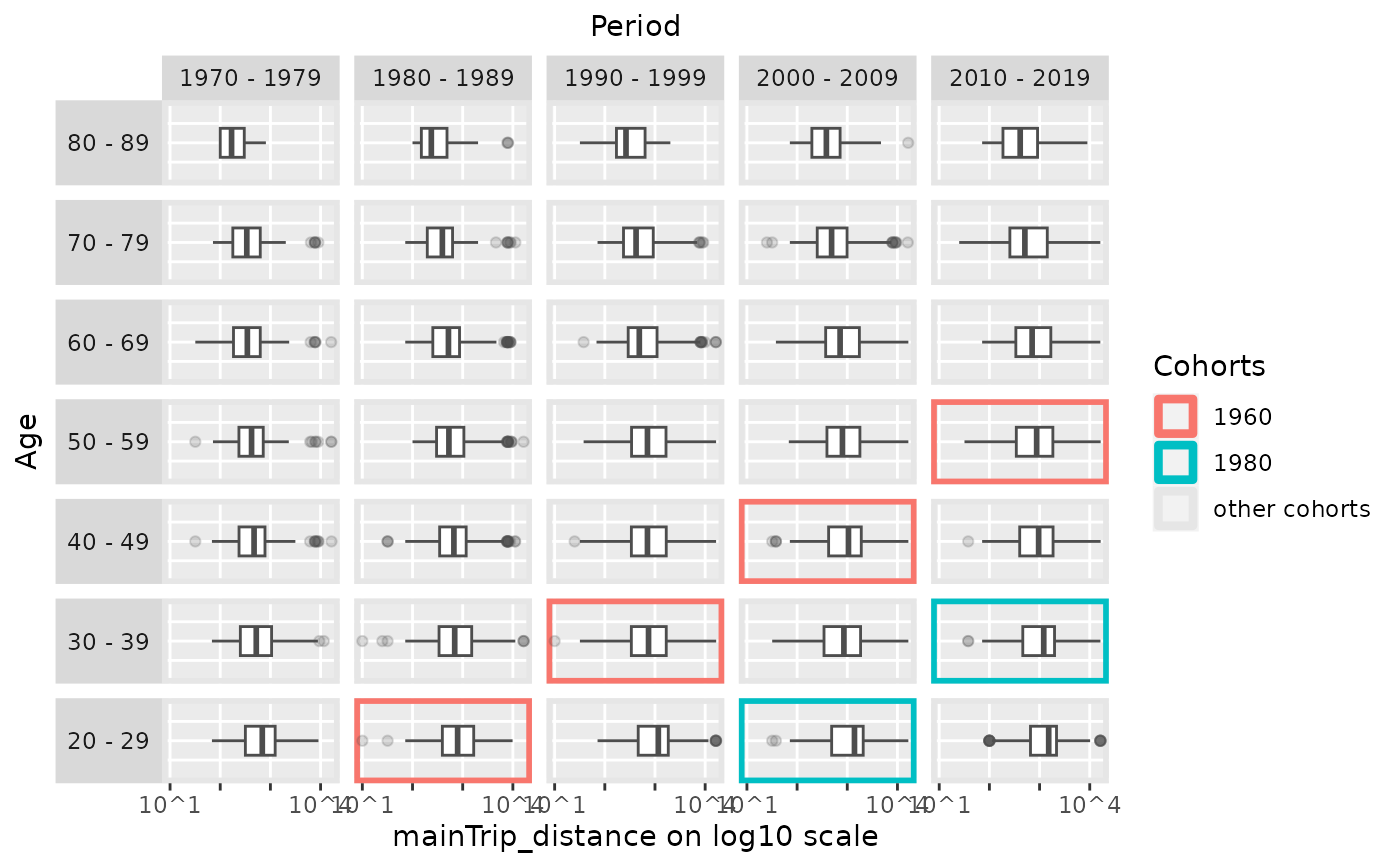
 # use boxplots instead of densities
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
plot_type = "boxplot",
age_groups = age_groups,
period_groups = period_groups,
log_scale = TRUE,
highlight_diagonals = list(8, 10))
#> Excluding 9149 missing observations of mainTrip_distance...
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
# use boxplots instead of densities
plot_densityMatrix(dat = travel,
y_var = "mainTrip_distance",
plot_type = "boxplot",
age_groups = age_groups,
period_groups = period_groups,
log_scale = TRUE,
highlight_diagonals = list(8, 10))
#> Excluding 9149 missing observations of mainTrip_distance...
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
#> Warning: Solution may be nonunique
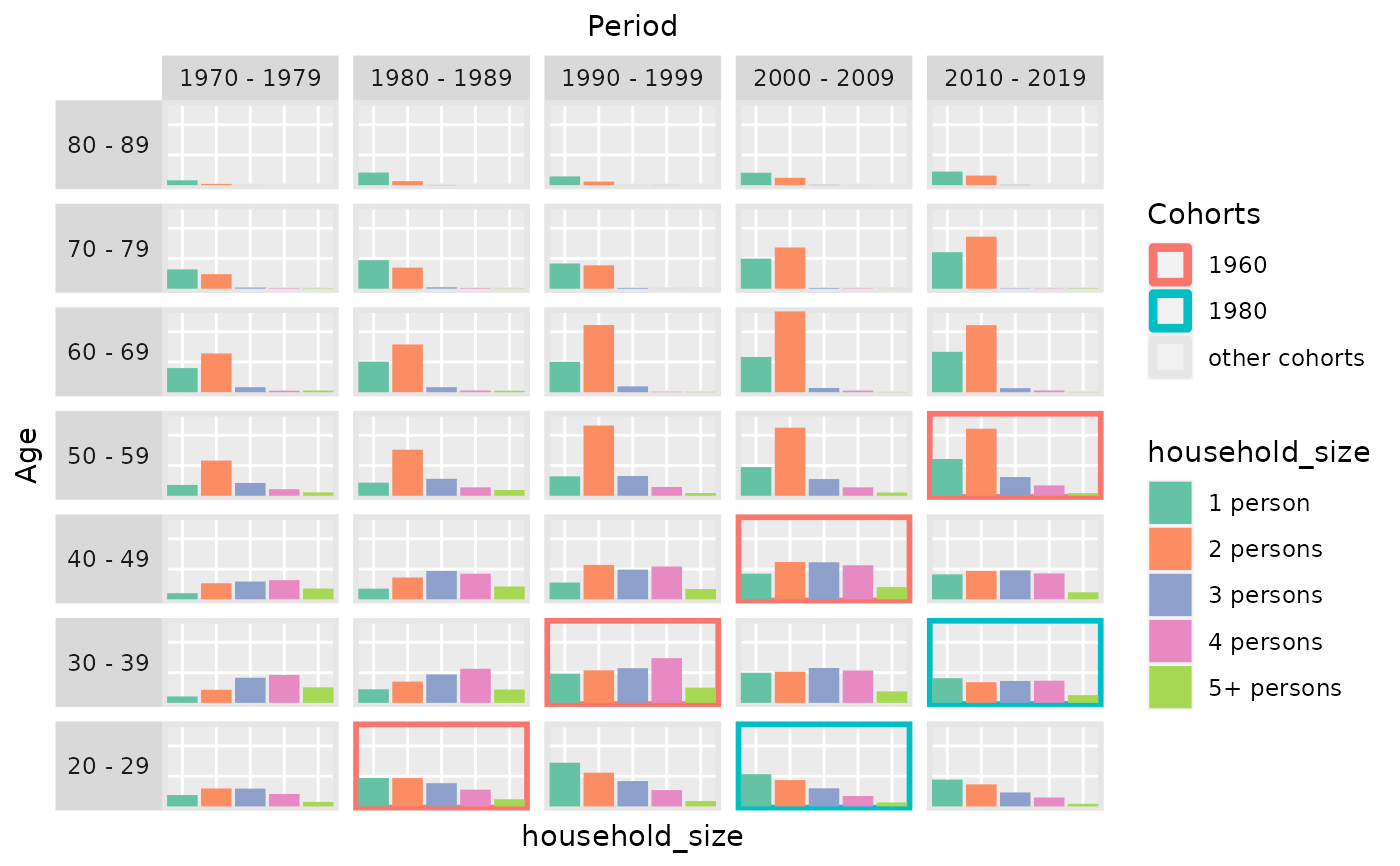
 # plot categorical variables instead of metric ones
plot_densityMatrix(dat = travel,
y_var = "household_size",
age_groups = age_groups,
period_groups = period_groups,
highlight_diagonals = list(8, 10))
# plot categorical variables instead of metric ones
plot_densityMatrix(dat = travel,
y_var = "household_size",
age_groups = age_groups,
period_groups = period_groups,
highlight_diagonals = list(8, 10))
 # }
# }